Guts UK is the charity for the digestive system. It funds scientific research such as Nicholas Ilott’s investigation into the link between the microbiome and chronic liver disorders, as Mewburn Ellis discovers.
Forward: features are independent pieces written for Mewburn Ellis discussing and celebrating the best of innovation and exploration from the scientific and entrepreneurial worlds.
Every morning Nicholas Ilott gets on his bike to cycle the short trip to the Kennedy Institute of Rheumatology at the University of Oxford, where he is a senior researcher working on several projects to understand how microbes in the human gut affect the liver and how this relates to disease.
His main research goal is to understand how the gut microbiome changes in individuals with primary sclerosing cholangitis (PSC). This is a chronic liver condition marked by scarring in the bile ducts, which can eventually lead to liver failure. But the liver does not act alone. ‘In liver diseases such as PSC, the liver impacts the microbiome,’ says Ilott.
He explains that reduced flow of bile to the intestine changes the microbiome and this affects the way that nutrients are absorbed. But molecules produced in the gut as a result of changes in the microbiome also feed back to the liver again. ‘It is this big inter-organ cycle, but we don't really understand all of the factors yet.’
Over the last few years research groups around the world have been studying the connection between the microbiome and PSC to find out, for example, how changes in the microbiome are related to the fact that many people with PSC also develop inflammatory bowel disease (IBD). Ilott is interested in learning how the intestinal microbiome affects the liver. ‘The most important question is whether we can target the intestine to help treat liver disease, because liver disease is the mortality-driving factor in PSC.’
Ilott’s research on the role of the microbiome in PSC is funded by an Early Career Researcher Grant from Guts UK. This charity supports patients with information and networks and funds researchers working on the digestive system through several types of grants, such as the one it awarded to Ilott. ‘The seed awards [it provides] are really crucial for giving early career researchers a jumpstart towards their own research,’ he says.
He is also involved in another project related to the gut liver axis. Together with a PhD student, Ilott is trying to understand more broadly how microbes control gene transcription in the liver and intestine. They study how genes are expressed within these organs in germ-free in vivo models, which don’t have a microbiome, compared with conventional models. This gives them an overview of which genes are controlled by the microbiome, which they can fine-tune to learn how individual strains of microbes are involved.
The intestinal microbiome:
Ilott is interested in learning how the intestinal microbiome affects the liver
The two projects are not entirely unrelated. ‘What we find here can be tied back to what we find in the PSC patient cohorts,’ says Ilott. A better understanding of how the microbiome affects gene expression in the liver could open the door to treatments that rely on manipulating the microbiome.
Ilott didn’t always specialise in the microbiome. He initially started in human genetics, and did a PhD in molecular psychiatry at King’s College London, where he studied how genetics affects activity level and attention deficit disorder. ‘That was an eye-opening experience for me,’ he recalls. ‘It became clear that genetic variants tend to have very small effects on traits like attention and it was difficult for me to see how I was going to move this forward.’
His research introduced him to high-throughput gene expression assays and this made him realise that understanding how to analyse large data sets was going to be an important skill. Ilott applied for a three-year MRC Career Development Fellowship, which took him to Oxford to learn computational biology. ‘The scheme was for biologists to get a proper foundation in bioinformatics and computational biology,’ he says. ‘After finishing that I had a nice mix of a very biological background with skills in computational biology.’
Around this time, Ilott first attended a talk about the microbiome and it piqued his interest. ‘I was just fascinated by the fact that you can tell what’s in people’s guts by sequencing.’ This inspired him to reach out to Professor Dame Fiona Powrie, who is currently director of the Kennedy Institute of Rheumatology, and he has been working in her research group ever since. Initially they collaborated on a project to investigate the genome and transcriptome of the microbiome in a model of inflammatory bowel disease, but now Ilott’s work has shifted to his independent research on the gut-liver axis.
Coming from a background in human genetics, studying the genetics of the microbiome was a bit of a shift for Ilott. ‘It’s a lot harder to study bacteria, because they’re essentially little metabolic factories.’ The metabolites that bacteria produce act on the human gut once they’re released there, so studying this ecosystem requires not only genomics but also metabolomics. Another challenge is that mutations occur very rapidly because microbes reproduce so quickly. ‘You see evolution in real time, which is something that we obviously don’t see in humans.’
Ilott uses a range of techniques in his work, including RNA sequencing to study bacterial transcription. To identify which microbes are present in samples from large patient cohorts he turns to an established method that involves reading a short sequence of 16S ribosomal RNA. This gene is present in all bacteria, but different between each type, which makes it a good marker of bacterial identity. For smaller sample sets he uses shotgun metagenomics, which reads all genetic material in a microbe sample. In addition to genetic analyses, Ilott also uses techniques such as ion chromatography, mass spectrometry and nuclear magnetic resonance to detect the metabolites produced by the microbiome.
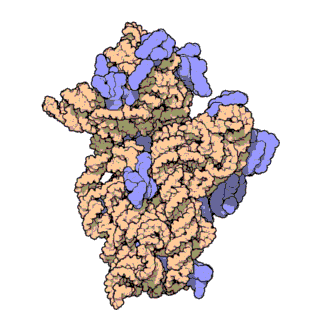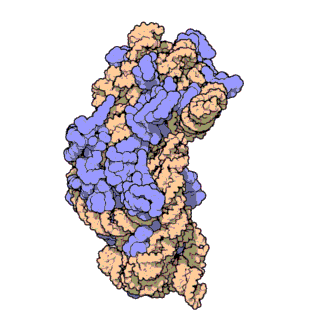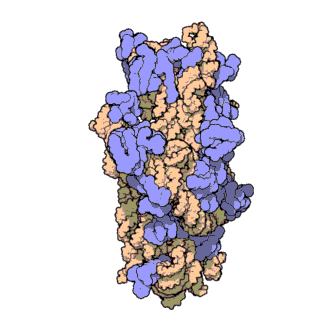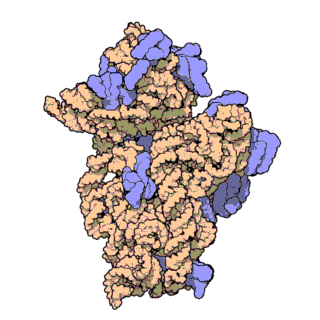
16S ribosomal RNA:
This gene is present in all bacteria, but different between each type, which makes it a good marker of bacterial identity
(Source: RCSB Protein Data Bank)
Even though existing techniques can reveal which microbes are present in a stool sample, the reality is that different microbes may live and act in different locations in the gut. That kind of location information would be very useful to capture as well. ‘There are new techniques for sampling the metabolome across the intestinal tract, using capsules that open at different pH along different parts of the gastrointestinal tract,’ says Ilott. ‘I think this is going to be really important for understanding complicated diseases where information is not all represented in the final output of the body.’ Another way to learn more about the location of microbes is by using spatial transcriptomics to see which cells specific microbes interact with in prepared tissue samples of the intestine.
In addition to his own research, Ilott is also involved with the Oxford Centre for Microbiome Studies (OCMS) as bioinformatics lead. OCMS facilitates microbiome research for scientists working at different departments across the University of Oxford by helping them with sample collection or study design. ‘It’s intended to give people access to the technology,’ Ilott says. ‘Particularly bioinformatics, because even if you’ve done RNA sequencing before, studying the microbiome is very different.’ As an example, he mentions that once you go down to species or strain level, the genetics of every bacterium is unique. However, different strains or species of bacteria have overlapping functions in the microbiome. This means that in the absence of one strain of bacteria another can take on its function. ‘This is why I think things like metabolomics and more functional outputs of the microbiome are important in addition to genomics,’ Ilott summarises.
What does this mean in practice? Is it possible yet to create personalised microbiome signatures to get information about medication response of disease progression? ‘I don’t think we’re quite there yet,’ says Ilott. ‘At the moment we don’t understand enough about the details of what bacteria do or the metabolites they produce.’ That makes it difficult to predict how microbiome data could be used in a clinical setting. Ilott does think that eventually this type of knowledge may inform tailored diets to adjust the microbiome, but he adds: ‘I think we need to understand more about the biology first.’
And to do that bioinformatics continues to play a central role. ‘Something that I think is really interesting is the marrying of classical microbiology with bioinformatics,’ muses Ilott. As an example he explains how it’s possible to create libraries of bacteria with different genetic modifications and grow these under conditions where some mutants survive and others don’t. With genetic sequencing and bioinformatics you can then track which mutated genes were linked to survival. ‘Even if we don’t know the function of the genes, we can very quickly and in a high-throughput way determine which genes are important for growth and survival under certain conditions,’ he says.
Ilott’s days are not all about bioinformatics. After his busy mornings working on different research projects and supporting the OCMS, he cycles home to spend the afternoon with his children. He is also an artist, mainly drawing portraits. ‘It allows me to switch off from everything,’ he says. Ilott creates digital science illustrations as well and has made some images for a Guts UK information leaflet about microbiome sampling.
In the next few years Ilott hopes to set up his own research group so he can continue his work on the gut-liver axis. That would allow him to address more of the unanswered questions that still remain in this field. ‘There’s a lot more to understand about the microbiome.’
Protecting microbiome based inventions and data driven technologies
Eliot Ward, Mewburn Ellis Partner and Patent Attorney, comments:
Nick’s insights into the functional relationships and redundancies in the microbiome raise important questions for those seeking to protect microbiome based inventions. If distinct microbial species can be substituted while maintaining the essential characteristics of a system, then functional definitions of the species become very important. Our microbiome team at Mewburn is adept at crafting suitable definitions to give our clients the best chance of obtaining meaningful protection for their valuable inventions.
Camille Terfve, Mewburn Ellis Partner and Patent Attorney, comments:
Computational biology plays an increasingly large role in all fields of modern biology. In microbiology in particular high throughput sequencing technologies were a paradigm shift as they enabled deep characterisation of the complexity of microbiomes, which typically contain a great variety of strains that can’t necessarily be grown in the lab. Protecting IP that is inherently linked to data driven technologies has its quirks as computer implemented inventions (patent jargon for solutions that are implemented at least partially in software) are examined differently from devices and compositions of matter. Our computational biology team at Mewburn specialises in dealing with those quirks to obtain strong IP for these inventions around the world, driven by extensive experience and a passion for data driven science.
Written by Eva Amsen
Eliot is a highly valued partner at Mewburn Ellis and a key driver within the Life Sciences team. With a reputation for driving complex projects from inception to completion, he works with a diverse range of clients, from innovative startups to multinational corporations worldwide. Eliot handles a diverse client portfolio spanning the life sciences and MedTech sectors. A skilled patent prosecutor, Eliot also has wide experience of drafting patent applications on breakthrough technologies, as well as leading offensive and defensive opposition proceedings post-grant. Eliot is also experienced in handling Freedom to Operate projects and in performing due diligence, which have led to the successful completion of high value transactions and investment rounds.
Email: eliot.ward@mewburn.com


-1.png)